import distl
import numpy as np
Multivariate Gaussian
First we'll create a multivariate gaussian distribution by providing the means and covariances of three parameters.
mvg = distl.mvgaussian([5,10, 12],
np.array([[ 2, 1, -1],
[ 1, 2, 1],
[-1, 1, 2]]),
allow_singular=True,
labels=['a', 'b', 'c'])
mvg.sample()
array([ 7.99080238, 11.46448835, 10.47368597])
mvg.sample(size=5)
array([[ 4.52006299, 10.4107841 , 12.89072111],
[ 4.6485998 , 9.62679128, 11.97819148],
[ 3.36823516, 9.32211349, 12.95387833],
[ 6.51815561, 12.77600097, 13.25784536],
[ 2.61171535, 6.80428169, 11.19256635]])
and plotting will now show a corner plot (if corner is installed)
fig = mvg.plot(show=True)
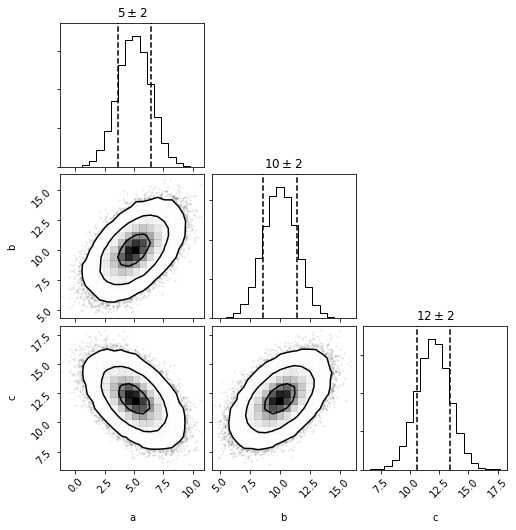
Multivariate Histogram
we can now convert this multivariate gaussian distribution into a multivariate histogram distribution (alternatively we could create a histogram directly from a set of samples or chains via mvhistogram_from_data.
mvh = mvg.to_mvhistogram(bins=15)
fig = mvh.plot(show=True, size=1e6)
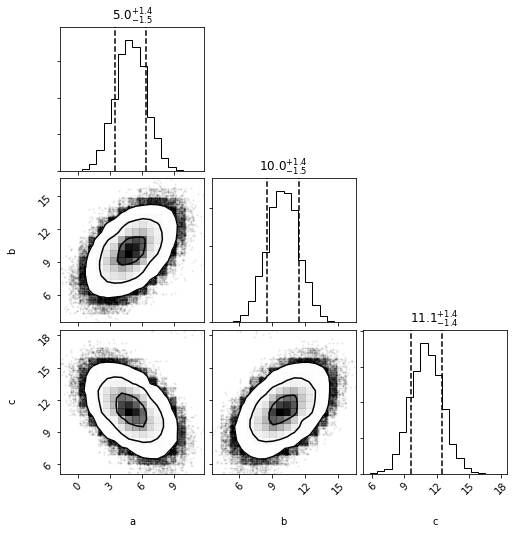
np.asarray(mvh.density.shape)
array([15, 15, 15])
Now if we access the means and covariances, we'll see that they are slightly different due to the binning.
mvh.calculate_means()
array([ 4.98034068, 9.97330153, 11.06628499])
mvh.calculate_covariances()
array([[ 2.14195908, 0.99110644, -1.00913987],
[ 0.99110644, 2.12047209, 0.99845476],
[-1.00913987, 0.99845476, 2.14286097]])
If we convert back to a multivariate gaussian, these are the means and covariances that will be adopted (technically not exactly as they'll be recomputed from another sampling of the underlying distribution).
mvhg = mvh.to_mvgaussian()
fig = mvhg.plot(show=True)
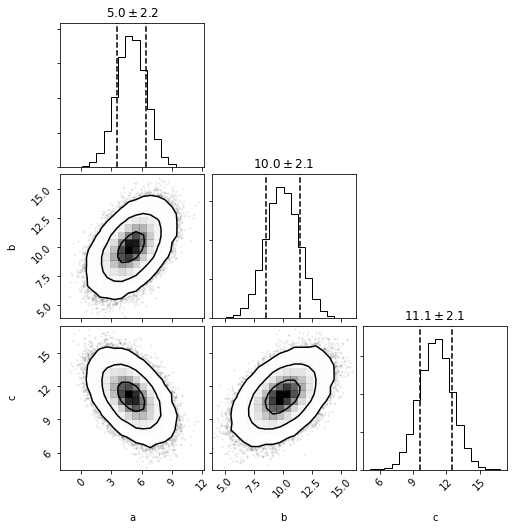
mvhg.mean
array([ 4.96639469, 9.9626855 , 11.05921629])
mvhg.cov
array([[ 2.15170257, 0.99878184, -1.01242674],
[ 0.99878184, 2.12055321, 0.9920415 ],
[-1.01242674, 0.9920415 , 2.14125743]])
Take Dimensions
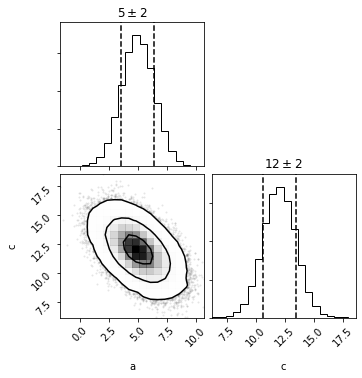
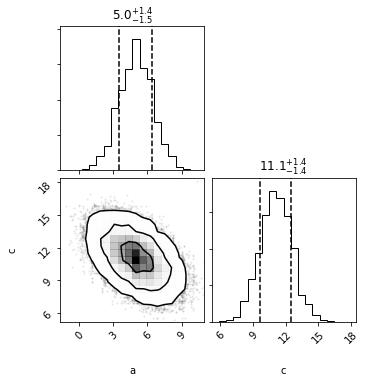
mvg_ac = mvg.take_dimensions(['a', 'c'])
mvg_ac.sample()
array([ 6.01363707, 13.52263276])
out = mvg_ac.plot(show=True)
out = mvh.take_dimensions(['a', 'c']).plot(show=True)
Passing a single dimension to take_dimension
If you pass a single-dimension to take_dimension, then the univariate version of the same type is returned instead. See the "Converting to Univariate" section below for examples directly calling to_univariate.
out = mvg.take_dimensions(['a']).plot(show=True)
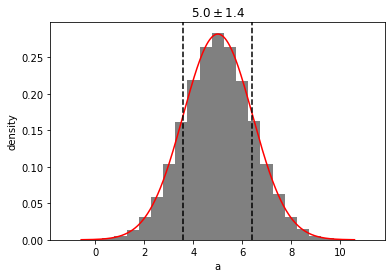
Slicing
Slicing allows taking a single dimension while retaining all underlying covariances such that the resulting distribution can undergo math operations, and/or logic, and included in distribution collections. For more details, see the slice examples.
mvg_a = mvg.slice('a')
mvg_a.sample()
6.160840301541623
out = mvg_a.plot(show=True)
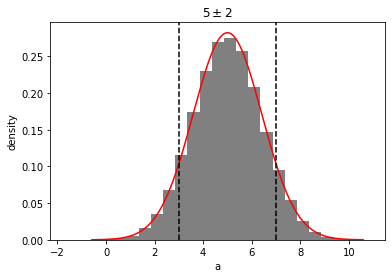
mvg_a.multivariate
<distl.mvgaussian mean=[5, 10, 12] cov=[[ 2 1 -1]
[ 1 2 1]
[-1 1 2]] allow_singular=True labels=['a', 'b', 'c']>
Converting to Univariate
There are methods to convert directly to the univariate distribution of the same type as the univariate:
When acting on a Multivariate, the requested dimension must be passed.
mvg.to_univariate(dimension='a')
<distl.gaussian loc=5.0 scale=1.4142135623730951 label=a>
Whereas a MultivariateSlice converts using the sliced dimension
mvg_a.to_univariate()
<distl.gaussian loc=5.0 scale=1.4142135623730951 label=a>